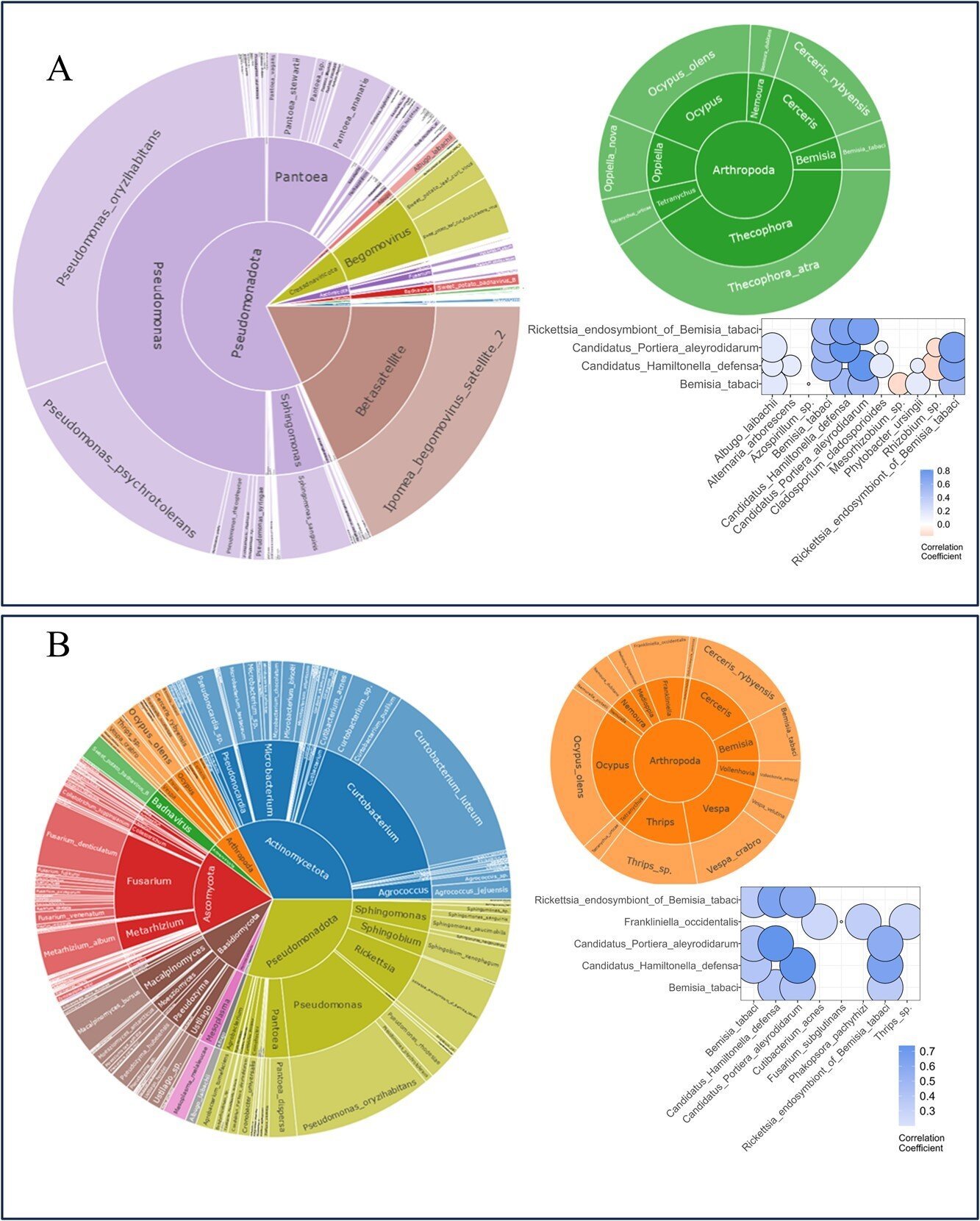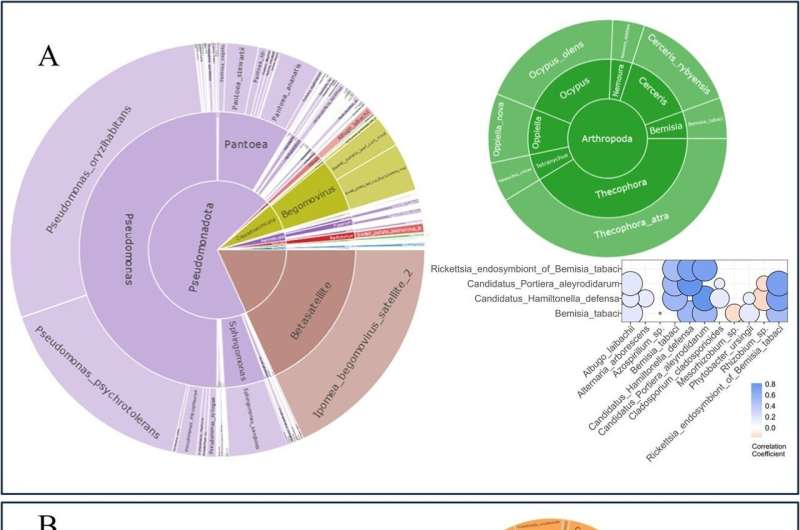
Sweet potato, a staple in combating global hunger, faces significant threats from pests like whiteflies and weevils, impacting plant growth and yields. A new study harnesses the power of genomic and metagenomic data to predict pest abundance and identify key genes that could fortify the plant’s defense mechanisms.
Sweet potatoes suffer significant yield losses due to pests like whiteflies and weevils, which affect plant growth and productivity. Traditional breeding methods face challenges in quickly addressing these complex pest interactions.
The role of microbial communities in plant defense is gaining attention as a potential solution. Due to these issues, there is an increasing need for in-depth research to understand and harness plant-microbiome interactions for developing pest-resistant sweet potato varieties.
Researchers from the University of Tennessee and their collaborators published a study on May 10, 2024, in Horticulture Research. The study investigates how metagenomic data can be used to enhance the understanding of plant-insect interactions in sweet potatoes. By integrating metagenome data, the researchers aim to improve the accuracy of genomic predictions for pest resistance.
The study utilized high-throughput sequencing and metagenome profiling to examine the interactions between sweet potatoes and insect pests, particularly whiteflies. Through quantitative reduced representation sequencing (qRRS), the researchers identified significant correlations between insect pests and various microbial communities within the sweet potato metagenome.
Notably, the study found that ethylene and cell wall modification pathways are crucial for resistance to whiteflies. By incorporating metagenome data as covariates in genomic prediction models, the researchers achieved a significant improvement in predictive accuracy for pest resistance.
This comprehensive approach revealed that modeling the metagenome alongside the host genome provides a more accurate prediction of pest resistance, emphasizing the potential of metagenome-informed breeding strategies to develop sweet potato varieties with enhanced pest resistance.
Dr. Bode A. Olukolu, the lead researcher, stated, “Our findings support the holobiont theory, suggesting that considering the metagenome alongside the host genome provides a more accurate prediction of pest resistance. This approach has the potential to revolutionize breeding strategies for sweet potatoes and other crops.”
The integration of metagenome data into breeding programs could lead to the development of sweet potato varieties with enhanced resistance to pests, reducing the reliance on chemical pesticides. This study paves the way for further research into the role of plant-associated microbial communities in crop protection and sustainability.
More information:
Alhagie K Cham et al, Metagenome-enabled models improve genomic predictive ability and identification of herbivory-limiting genes in sweetpotato, Horticulture Research (2024). DOI: 10.1093/hr/uhae135
Provided by
NanJing Agricultural University
Citation:
Battling bugs with big data: Sweet potato’s genomic-metagenomic pest shield (2024, August 8)
retrieved 8 August 2024
from https://phys.org/news/2024-08-bugs-big-sweet-potato-genomic.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.